
NMR Restraints Grid
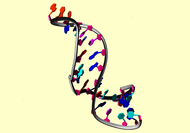
 |
NMR Restraints Grid |
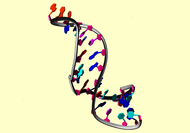 |
Result table
| image | mrblock_id | pdb_id | bmrb_id | cing | stage | position | program | type | subtype | subsubtype |
|
|
8045 |
1ldz |
4226 | cing | 1-original | 3 | XPLOR/CNS | distance | general distance | simple |
! ! 6. G imino to H1' NOEs (indirect distances involving NH2 group: ! 1.8 - 5.0 Angstrom for residues in helices ! formed by 1-4 11-14, 19-22, 27-30 ! 1.8 - 6.8 Angstrom for others ) ! ! 1.8 - 6 A H1' to G amino nitrogen ! assign (resid 26 and name n2 ) (resid 27 and name h1' ) 3.90 3.90 2.10 assign (resid 1 and name n2 ) (resid 2 and name h1' ) 3.90 3.90 2.10 assign (resid 19 and name n2 ) (resid 20 and name h1' ) 3.90 3.90 2.10 assign (resid 19 and name n2 ) (resid 15 and name h1' ) 3.90 3.90 2.60 assign (resid 29 and name n2 ) (resid 30 and name h1' ) 3.90 3.90 2.10 assign (resid 29 and name n2 ) (resid 3 and name h1' ) 3.90 3.90 2.10 assign (resid 23 and name n2 ) (resid 11 and name h1' ) 3.90 3.90 2.10 assign (resid 3 and name n2 ) (resid 4 and name h1' ) 3.90 3.90 2.10 assign (resid 3 and name n2 ) (resid 29 and name h1' ) 3.90 3.90 2.10 assign (resid 22 and name n2 ) (resid 23 and name h1' ) 3.90 3.90 2.10 assign (resid 22 and name n2 ) (resid 12 and name h1' ) 3.90 3.90 2.10 assign (resid 13 and name n2 ) (resid 14 and name h1' ) 3.90 3.90 2.10 assign (resid 13 and name n2 ) (resid 21 and name h1' ) 3.90 3.90 2.10 ! ! ! ! 7. G/U imino to other NH2s NOEs (direct distances: 1.8 - 7.0 Angstrom ) ! ! imino to amino nitrogen 1.8 - 6 A ! ! assign (resid 14 and name n4 ) (resid 20 and name h3 ) 3.90 3.90 2.10 assign (resid 14 and name n4 ) (resid 13 and name h1 ) 3.90 3.90 2.10 assign (resid 3 and name n2 ) (resid 27 and name h3 ) 3.90 3.90 2.10 assign (resid 5 and name n4 ) (resid 27 and name h3 ) 3.90 3.90 2.10 assign (resid 11 and name n4 ) (resid 21 and name h3 ) 3.90 3.90 2.10 assign (resid 28 and name n4 ) (resid 29 and name h1 ) 3.90 3.90 2.10 ! ! ! ! 8. Aromatics to NH NOEs (direct distances: 1.8 - 7.0 Angstrom ! ! ! ! 9. Others ! assign (resid 25 and name H2 ) (resid 5 and name N4 ) 3.90 3.90 2.10 ! ! ! file lz2_ami2.noe 5/07/95 ! ! This file contains the distances derived from the observed NOE ! in Luci's spectrum: leadz_cpmg-xy_noe-16c-2.mat and leadz_cpmg-xy_noe_24c.mat ! and which have not been observed in other NOESY spectra ! ref. Mueller et al. JACS 117, 11043 (1995) ! ! NOESY to amino are defined to the nitrogen ! ! The distance is defined as: ! 1.8 to 6.0 A involving one amino ! 1.8 to 7.0 A involving two aminos ! except for A18NH2 to G15 H1' (1.8 to 4.0) ! ! The distance is redefined as: (5/7/95) ! 1.8 to 7.0 A involving one amino ! 1.8 to 8.0 A involving two aminos ! except for A18NH2 to G15 H1' (1.8 to 4.0) ! ! Modified CGH 071195 reset A18
Please acknowledge these references in publications where the data from this site have been utilized.Contact the webmaster for help, if required. Wednesday, May 1, 2024 1:53:22 PM GMT (wattos1)