
NMR Restraints Grid
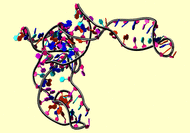
 |
NMR Restraints Grid |
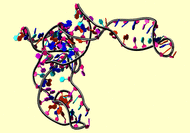 |
Result table
| image | mrblock_id | pdb_id | cing | stage | position | program | type | subtype | subsubtype |
|
|
497572 |
2krl |
cing | 1-original | 4 | XPLOR/CNS | distance | general distance | simple |
!===================================================
! Global restraints from SAXS molecular envelope
!===================================================
assign
(( resid 10 and name P ))
(( resid 101 and name P ))
64.0 2.00 2.00
assign
(( resid 11 and name P ))
(( resid 101 and name P ))
64.0 2.00 2.00
assign
(( resid 35 and name P ))
(( resid 77 and name P ))
80.0 2.00 2.00
assign
(( resid 1 and name C3' ))
(( resid 53 and name P ))
5.0 2.00 2.00
assign
(( resid 20 and name P ))
(( resid 27 and name P ))
7.0 1.00 1.00
assign
(( resid 10 and name P ))
(( resid 35 and name P ))
55.0 5.00 5.00
assign
(( resid 10 and name P ))
(( resid 45 and name P ))
55.0 5.00 5.00
assign
(( resid 11 and name P ))
(( resid 35 and name P ))
55.0 5.00 5.00
assign
(( resid 11 and name P ))
(( resid 45 and name P ))
55.0 5.00 5.00
assign
(( resid 50 and name P ))
(( resid 59 and name P ))
33.0 5.00 5.00
assign
(( resid 50 and name P ))
(( resid 93 and name P ))
33.0 5.00 5.00
assign
(( resid 50 and name P ))
(( resid 96 and name P ))
30.0 5.00 5.00
assign
(( resid 63 and name P ))
(( resid 87 and name P ))
16.0 5.00 5.00
assign
(( resid 62 and name P ))
(( resid 87 and name P ))
16.0 5.00 5.00
!---------------------------------------------
! local flexible duplex restraints
!---------------------------------------------
assign
(( resid 67 and name P ))
(( resid 68 and name P ))
5.4 0.500 0.5000
assign
(( resid 68 and name P ))
(( resid 69 and name P ))
5.4 0.500 0.5000
assign
(( resid 69 and name P ))
(( resid 70 and name P ))
5.4 0.500 0.5000
assign
(( resid 70 and name P ))
(( resid 71 and name P ))
5.4 0.500 0.5000
assign
(( resid 71 and name P ))
(( resid 72 and name P ))
5.4 0.500 0.5000
assign
(( resid 72 and name P ))
(( resid 73 and name P ))
5.4 0.500 0.5000
assign
(( resid 82 and name P ))
(( resid 83 and name P ))
5.4 0.500 0.5000
ssign
(( resid 83 and name P ))
(( resid 84 and name P ))
5.4 0.500 0.5000
assign
(( resid 84 and name P ))
(( resid 85 and name P ))
5.4 0.500 0.5000
assign
(( resid 85 and name P ))
(( resid 86 and name P ))
5.4 0.500 0.5000
assign
(( resid 86 and name P ))
(( resid 87 and name P ))
5.4 0.500 0.5000
assign
(( resid 87 and name P ))
(( resid 88 and name P ))
5.4 0.500 0.5000
assign
(( resid 67 and name P ))
(( resid 88 and name P ))
18.0 0.500 2.000
assign
(( resid 68 and name P ))
(( resid 87 and name P ))
18.0 0.500 2.000
assign
(( resid 70 and name P ))
(( resid 85 and name P ))
18.0 0.500 2.000
assign
(( resid 71 and name P ))
(( resid 84 and name P ))
18.0 0.500 2.000
assign
(( resid 72 and name P ))
(( resid 83 and name P ))
18.0 0.500 2.000
Contact the webmaster for help, if required. Sunday, April 28, 2024 12:24:35 AM GMT (wattos1)