
NMR Restraints Grid
 |
NMR Restraints Grid |
 |
| File name | Description |
| readme.html | This file |
| index.csv | Index file used for looking up mr_block_id(s) for specific entries or data types |
| main.css | Hypertext Cascading Style Sheet used by your browser for rendering the readme. Keep this file in the same directory as the readme. Not important otherwise. |
| errors.txt | An optional file that describes any errors that might have been encountered downloading the data from the database. It is possible that modifications to the database during download removed requested blocks. That is one example that would be documented in this file. |
| block_123.txt | File with data on the block with mr_block_id equal to e.g. 123 |
| block_124.tgz | File with binary data of a gzipped tar file with for example XML files. A full list of possible file extensions is detailed below. |
| block_125.txt | Etc. |
gawk -F',' '{if($4=="XXXX")print $2}' index.csv > block_ids.txt
Note that the field number of the PDB code might be 3 in stead of 4 depending on whether or not any BMRB codes are listed.| Filename extension | Explanation |
| aco | DYANA/DIANA for dihedral angle restraints |
| csv | Comma Separate Values |
| lol | DYANA/DIANA upper limits for distance restraints |
| mtf | CNS Molecular Topology File |
| pdb | Protein DataBank |
| seq | DYANA/DIANA sequence |
| str | STAR: Self-defining Text Archive and Retrieval |
| tbl | XPLOR/CNS tables of data |
| tgz | Gzipped tar |
| txt | General text |
| upl | DYANA/DIANA upper limits for distance restraints |
| xml | EXtensible Markup Language |
| Column | Explanation |
| bmrb_id | BMRB accession number |
| format | Format of the data e.g. "ambi" for ambiguous distance restraints |
| image | Molecular image of the PDB entry |
| in_dress | True if the structure has been recalculated in DRESS |
| in_recoord | True if the structure has been recalculated in RECOORD |
| item_count | Total number of distance, dihedral angle, and RDC restraints. Currently only STAR formatted files generated at BMRB are counted. |
| mrblock_id | Numeric id in the database for this block (may change over time) |
| mrfile_id | Numeric id in the database for the file this block is part of |
| other_prop | Other type of information e.g. "LOWER_ONLY=true" for CYANA data |
| pdb_id | PDB code being a numeral followed by three alphanumericals |
| position | Position of this block in the file (the first block is numbered 0) |
| program | Program or software format, e.g. XPLOR/CNS, STAR |
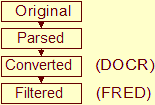
| stage | Data flows from original, parsed, converted, to filtered, see the NMR Restraint Grid paper |
| subtype | Subtype of the data such as "NOE" for distance restraints |
| type | Type of the data such as "distance" for distance restraints |